Importing ARCOS Data with Dask#
Last week, we used dask to play with a few datasets to get a feel for how dask works. In order to help us develop code that would run quickly, however, we worked with very small, safe datasets.
Today, we will continue to work with dask, but this time using much larger datasets. This means that (a) doing things incorrectly may lead to your computer crashing (So save all your open files before you start!), and (b) many of the commands you are being asked run will take several minutes each.
For familiarity, and so you can see what advantages dask can bring to your workflow, today we’ll be working with the DEA ARCOS drug shipment database published by the Washington Post! However, to strike a balance between size and speed, we’ll be working with a slightly thinned version that has only the last two years of data, instead of all six.
Exercise 1#
Download the thinned ARCOS data from this link. It should be about 2GB zipped, 25 GB unzipped.
Exercise 2#
Our goal today is going to be to find the pharmaceutical company that has shipped the most opioids (MME_Conversion_Factor * CALC_BASE_WT_IN_GM) in the US.
When working with large datasets, it is good practice to begin by prototyping your code with a subset of your data. So begin by using pandas to read in the first 100,000 lines of the ARCOS data and write pandas code to compute the shipments from each shipper (the group that reported the shipment).
import pandas as pd
import numpy as np
import os
pd.set_option("mode.copy_on_write", True)
os.chdir("/users/nce8/downloads/")
df = pd.read_csv("arcos_2011_2012.tsv", nrows=100_000, sep="\t")
df.sample().T
/var/folders/fs/h_8_rwsn5hvg9mhp0txgc_s9v6191b/T/ipykernel_73426/2703016818.py:8: DtypeWarning: Columns (4,6,27) have mixed types. Specify dtype option on import or set low_memory=False.
df = pd.read_csv("arcos_2011_2012.tsv", nrows=100_000, sep="\t")
| 51243 | |
|---|---|
| Unnamed: 0 | 68243 |
| REPORTER_DEA_NO | PL0032627 |
| REPORTER_BUS_ACT | DISTRIBUTOR |
| REPORTER_NAME | AMERISOURCEBERGEN DRUG CORP |
| REPORTER_ADDL_CO_INFO | NaN |
| REPORTER_ADDRESS1 | 322 N 3RD STREET |
| REPORTER_ADDRESS2 | NaN |
| REPORTER_CITY | PADUCAH |
| REPORTER_STATE | KY |
| REPORTER_ZIP | 42001 |
| REPORTER_COUNTY | MCCRACKEN |
| BUYER_DEA_NO | BF1167027 |
| BUYER_BUS_ACT | RETAIL PHARMACY |
| BUYER_NAME | FRED'S PHARMACY |
| BUYER_ADDL_CO_INFO | NaN |
| BUYER_ADDRESS1 | 475 HIGHWAY 6 EAST |
| BUYER_ADDRESS2 | NaN |
| BUYER_CITY | BATESVILLE |
| BUYER_STATE | MS |
| BUYER_ZIP | 38606 |
| BUYER_COUNTY | PANOLA |
| TRANSACTION_CODE | S |
| DRUG_CODE | 9193 |
| NDC_NO | 591054005 |
| DRUG_NAME | HYDROCODONE |
| QUANTITY | 1.0 |
| UNIT | NaN |
| ACTION_INDICATOR | NaN |
| ORDER_FORM_NO | NaN |
| CORRECTION_NO | NaN |
| STRENGTH | NaN |
| TRANSACTION_DATE | 1062012 |
| CALC_BASE_WT_IN_GM | 3.027 |
| DOSAGE_UNIT | 500.0 |
| TRANSACTION_ID | 21858 |
| Product_Name | HYDROCODONE BIT. 10MG/ACETAMINOPHEN |
| Ingredient_Name | HYDROCODONE BITARTRATE HEMIPENTAHYDRATE |
| Measure | TAB |
| MME_Conversion_Factor | 1.0 |
| Combined_Labeler_Name | Actavis Pharma, Inc. |
| Revised_Company_Name | Allergan, Inc. |
| Reporter_family | AmerisourceBergen Drug |
| dos_str | 10.0 |
| date | 2012-10-06 |
| year | 2012 |
df["mme"] = df["MME_Conversion_Factor"] * df["CALC_BASE_WT_IN_GM"]
grouped = df.groupby(["REPORTER_NAME"], as_index=False)["mme"].sum()
grouped.sort_values("mme", ascending=False)
| REPORTER_NAME | mme | |
|---|---|---|
| 20 | MCKESSON CORPORATION | 299266.331225 |
| 7 | CARDINAL HEALTH 110, LLC | 54352.323711 |
| 2 | AMERISOURCEBERGEN DRUG CORP | 34561.394892 |
| 17 | KINRAY INC | 28620.315246 |
| 19 | LOUISIANA WHOLESALE DRUG CO | 14787.765559 |
| 11 | FRANK W KERR INC | 8730.016283 |
| 12 | H D SMITH WHOLESALE DRUG CO | 6399.324050 |
| 16 | KAISER FOUNDATION HOSPITALS | 3891.329580 |
| 5 | BURLINGTON DRUG COMPANY | 3889.490325 |
| 1 | AMERICAN SALES COMPANY | 3432.058005 |
| 9 | DIK DRUG CO | 3278.514405 |
| 18 | KPH HEALTHCARE SERVICES, INC. | 1988.890350 |
| 4 | BLOODWORTH WHOLESALE DRUGS | 1782.827325 |
| 10 | DISCOUNT DRUG MART | 1272.596205 |
| 15 | KAISER FNDTN HEALTH PLAN NW | 1159.892040 |
| 13 | H J HARKINS COMPANY INC | 352.393956 |
| 6 | CAPITAL WHOLESALE DRUG & CO | 188.318175 |
| 3 | APOTHECA INC | 23.913300 |
| 14 | HALS MED DENT SUPPLY CO., INC. | 18.162000 |
| 8 | CESAR CASTILLO INC | 3.027000 |
| 21 | MERRITT VETERINARY SUPPLIES INC | 1.816200 |
| 0 | ACE SURGICAL SUPPLY CO INC | 0.605400 |
Exercise 3#
Now let’s turn to dask. Re-write your code for dask, and calculate the total shipments by reporting company. Remember:
Activate a conda environment with a clean dask installation.
Start by spinning up a distributed cluster.
Dask won’t read compressed files, so you have to unzip your ARCOS data.
Start your cluster in a cell all by itself since you don’t want to keep re-running the “start a cluster” code.
If you need to review dask basic code, check here.
As you run your code, make sure to click on the Dashboard link below where you created your cluster:
Among other things, the bar across the bottom should give you a sense of how long your task will take:
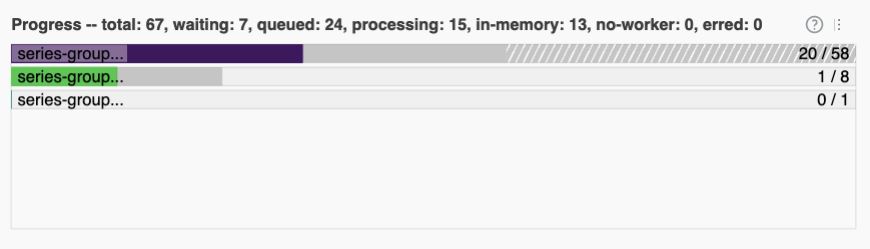
(For context, my computer (which has 10 cores) only took a couple seconds. My computer is fast, but most computers should be done within a couple minutes, tops).
from dask.distributed import Client
client = Client()
client
/Users/nce8/miniforge3/envs/dask/lib/python3.13/site-packages/distributed/node.py:187: UserWarning: Port 8787 is already in use.
Perhaps you already have a cluster running?
Hosting the HTTP server on port 62107 instead
warnings.warn(
Client
Client-e5c507f6-a6a0-11ef-9ed2-f623161a15ec
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:62107/status |
Cluster Info
LocalCluster
af36c3c4
| Dashboard: http://127.0.0.1:62107/status | Workers: 5 |
| Total threads: 10 | Total memory: 64.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-3656ae48-9b0e-4809-b57e-ff963558e872
| Comm: tcp://127.0.0.1:62108 | Workers: 5 |
| Dashboard: http://127.0.0.1:62107/status | Total threads: 10 |
| Started: Just now | Total memory: 64.00 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:62126 | Total threads: 2 |
| Dashboard: http://127.0.0.1:62127/status | Memory: 12.80 GiB |
| Nanny: tcp://127.0.0.1:62111 | |
| Local directory: /var/folders/fs/h_8_rwsn5hvg9mhp0txgc_s9v6191b/T/dask-scratch-space/worker-z9okim4v | |
Worker: 1
| Comm: tcp://127.0.0.1:62123 | Total threads: 2 |
| Dashboard: http://127.0.0.1:62124/status | Memory: 12.80 GiB |
| Nanny: tcp://127.0.0.1:62113 | |
| Local directory: /var/folders/fs/h_8_rwsn5hvg9mhp0txgc_s9v6191b/T/dask-scratch-space/worker-yk0ugh1n | |
Worker: 2
| Comm: tcp://127.0.0.1:62129 | Total threads: 2 |
| Dashboard: http://127.0.0.1:62130/status | Memory: 12.80 GiB |
| Nanny: tcp://127.0.0.1:62115 | |
| Local directory: /var/folders/fs/h_8_rwsn5hvg9mhp0txgc_s9v6191b/T/dask-scratch-space/worker-rcqhmlid | |
Worker: 3
| Comm: tcp://127.0.0.1:62132 | Total threads: 2 |
| Dashboard: http://127.0.0.1:62133/status | Memory: 12.80 GiB |
| Nanny: tcp://127.0.0.1:62117 | |
| Local directory: /var/folders/fs/h_8_rwsn5hvg9mhp0txgc_s9v6191b/T/dask-scratch-space/worker-okijjlir | |
Worker: 4
| Comm: tcp://127.0.0.1:62135 | Total threads: 2 |
| Dashboard: http://127.0.0.1:62136/status | Memory: 12.80 GiB |
| Nanny: tcp://127.0.0.1:62119 | |
| Local directory: /var/folders/fs/h_8_rwsn5hvg9mhp0txgc_s9v6191b/T/dask-scratch-space/worker-984ttoki | |
import dask.dataframe as dd
df = dd.read_csv(
"/users/nce8/downloads/arcos_2011_2012.tsv",
sep="\t",
usecols=["REPORTER_NAME", "MME_Conversion_Factor", "CALC_BASE_WT_IN_GM"],
)
df["mme"] = df["MME_Conversion_Factor"] * df["CALC_BASE_WT_IN_GM"]
grouped = df.groupby(["REPORTER_NAME"])["mme"].sum()
results = grouped.compute()
2024-11-19 13:05:46,592 - distributed.shuffle._scheduler_plugin - WARNING - Shuffle 598788feafe30f29ea9811cb7fb2caa2 initialized by task ('shuffle-transfer-598788feafe30f29ea9811cb7fb2caa2', 378) executed on worker tcp://127.0.0.1:62132
2024-11-19 13:06:02,885 - distributed.shuffle._scheduler_plugin - WARNING - Shuffle 598788feafe30f29ea9811cb7fb2caa2 deactivated due to stimulus 'task-finished-1732039562.883873'
2024-11-19 13:06:03,561 - distributed.shuffle._scheduler_plugin - WARNING - Shuffle 942382d9008028879f257963906f7205 initialized by task ('shuffle-transfer-942382d9008028879f257963906f7205', 99) executed on worker tcp://127.0.0.1:62129
2024-11-19 13:06:21,346 - distributed.shuffle._scheduler_plugin - WARNING - Shuffle 942382d9008028879f257963906f7205 deactivated due to stimulus 'task-finished-1732039581.3442008'
2024-11-19 13:06:22,008 - distributed.shuffle._scheduler_plugin - WARNING - Shuffle 36907e7bcfab7b9ba83c9c7885ff5208 initialized by task ('shuffle-transfer-36907e7bcfab7b9ba83c9c7885ff5208', 191) executed on worker tcp://127.0.0.1:62126
2024-11-19 13:06:42,666 - distributed.shuffle._scheduler_plugin - WARNING - Shuffle 36907e7bcfab7b9ba83c9c7885ff5208 deactivated due to stimulus 'task-finished-1732039602.665749'
results.sort_values(ascending=False) / 1_000
REPORTER_NAME
MCKESSON CORPORATION 56046.787034
CARDINAL HEALTH 46719.583860
WALGREEN CO 41850.332089
AMERISOURCEBERGEN DRUG CORP 25533.636319
CARDINAL HEALTH 110, LLC 5896.801450
...
QUIQ, INC 0.000533
SOUTHERN MEDICAL LASERS DBA SML MEDICAL SALES 0.000454
GAVIS PHARMACEUTICALS, LLC 0.000303
REMEDYREPACK 0.000182
MIKART 0.000170
Name: mme, Length: 316, dtype: float64
Exercise 4#
Now let’s calculate, for each state, what company shipped the most pills?
Note you will quickly find that you can’t sort in dask – sorting in parallel is really tricky! So you’ll have to work around that. Do what you need to do on the big dataset first, then compute it all so you get it as a regular pandas dataframe, then finish.
import dask.dataframe as dd
df = dd.read_csv(
"arcos_2011_2012.tsv",
sep="\t",
usecols=[
"REPORTER_NAME",
"MME_Conversion_Factor",
"CALC_BASE_WT_IN_GM",
"BUYER_STATE",
],
)
df["mme"] = df["MME_Conversion_Factor"] * df["CALC_BASE_WT_IN_GM"]
grouped = df.groupby(["REPORTER_NAME", "BUYER_STATE"])["mme"].sum().reset_index()
results = grouped.compute()
results = results.sort_values(["mme"], ascending=False)
state_maxes = results.groupby(["BUYER_STATE"]).nth(0)
state_maxes
| REPORTER_NAME | BUYER_STATE | mme | |
|---|---|---|---|
| 109 | WALGREEN CO | FL | 6.566340e+06 |
| 3 | MCKESSON CORPORATION | CA | 5.152963e+06 |
| 0 | CARDINAL HEALTH 110, LLC | NY | 4.014733e+06 |
| 2 | MCKESSON CORPORATION | PA | 3.637664e+06 |
| 118 | CARDINAL HEALTH | OH | 2.966956e+06 |
| 11 | WALGREEN CO | TX | 2.318831e+06 |
| 12 | WALGREEN CO | AZ | 2.312494e+06 |
| 6 | CARDINAL HEALTH | NC | 2.273949e+06 |
| 2 | MCKESSON CORPORATION | MD | 2.230794e+06 |
| 102 | MCKESSON CORPORATION | AL | 2.143844e+06 |
| 5 | MCKESSON CORPORATION | MI | 1.998488e+06 |
| 6 | CARDINAL HEALTH | TN | 1.943907e+06 |
| 13 | CARDINAL HEALTH | IN | 1.881065e+06 |
| 6 | MCKESSON CORPORATION | NJ | 1.807488e+06 |
| 7 | CARDINAL HEALTH | MA | 1.790407e+06 |
| 20 | WALGREEN CO | WI | 1.778450e+06 |
| 22 | WALGREEN CO | IL | 1.774219e+06 |
| 4 | MCKESSON CORPORATION | WA | 1.680772e+06 |
| 14 | MCKESSON CORPORATION | GA | 1.643461e+06 |
| 9 | CARDINAL HEALTH | VA | 1.530984e+06 |
| 27 | MCKESSON CORPORATION | SC | 1.372027e+06 |
| 5 | MCKESSON CORPORATION | OK | 1.369128e+06 |
| 3 | MCKESSON CORPORATION | MO | 1.331515e+06 |
| 80 | MCKESSON CORPORATION | OR | 1.297825e+06 |
| 97 | CARDINAL HEALTH | NV | 1.293392e+06 |
| 0 | AMERISOURCEBERGEN DRUG CORP | KY | 1.270813e+06 |
| 23 | WALGREEN CO | CO | 1.044110e+06 |
| 4 | CARDINAL HEALTH | CT | 1.016512e+06 |
| 31 | MORRIS & DICKSON CO | LA | 8.171550e+05 |
| 18 | CARDINAL HEALTH | WV | 7.574662e+05 |
| 149 | MCKESSON DRUG COMPANY | MN | 6.925496e+05 |
| 30 | MCKESSON CORPORATION | KS | 6.677067e+05 |
| 15 | AMERISOURCEBERGEN DRUG CORP | UT | 6.667854e+05 |
| 42 | WALGREEN CO | DE | 6.339375e+05 |
| 2 | MCKESSON CORPORATION | AR | 6.053744e+05 |
| 4 | CARDINAL HEALTH | ME | 5.288570e+05 |
| 105 | WALGREEN CO | NM | 5.234781e+05 |
| 74 | MCKESSON CORPORATION | MS | 5.128833e+05 |
| 3 | MCKESSON CORPORATION | NH | 3.731772e+05 |
| 3 | MCKESSON CORPORATION | MT | 3.608250e+05 |
| 133 | AMERISOURCEBERGEN DRUG | IA | 3.421264e+05 |
| 1 | MCKESSON CORPORATION | HI | 3.371645e+05 |
| 7 | CARDINAL HEALTH | RI | 3.344156e+05 |
| 1 | MCKESSON CORPORATION | ID | 3.213065e+05 |
| 0 | MCKESSON CORPORATION | NE | 2.415497e+05 |
| 4 | MCKESSON CORPORATION | VT | 1.693842e+05 |
| 3 | CARDINAL HEALTH | AK | 1.549137e+05 |
| 21 | CARDINAL HEALTH | DC | 1.329334e+05 |
| 48 | MCKESSON CORPORATION | SD | 1.206526e+05 |
| 5 | MCKESSON CORPORATION | WY | 1.016961e+05 |
| 58 | MCKESSON DRUG COMPANY | ND | 1.014997e+05 |
| 5 | DROGUERIA BETANCES, LLC | PR | 7.926534e+04 |
| 14 | AMERISOURCEBERGEN DRUG CORP | GU | 7.272706e+03 |
| 34 | CARDINAL HEALTH P.R. 120, INC. | VI | 3.696650e+03 |
| 112 | AMERISOURCEBERGEN DRUG CORP | MP | 2.194500e+03 |
Does this seem like a situation where a single company is responsible for the opioid epidemic?
Exercise 5#
Now go ahead and try and re-do the importation of ARCOS data you did by hand for your opioid analysis project (In other words, for each year, calculate the total morphine equivalents sent to each county in the US, but do it with this 2 years of data).
Rather than chunking by hand — as you should have done previously for the opioid analysis project — now let dask do all that hard work automatically.
import dask.dataframe as dd
df = dd.read_csv(
"arcos_2011_2012.tsv",
sep="\t",
usecols=[
"MME_Conversion_Factor",
"CALC_BASE_WT_IN_GM",
"BUYER_STATE",
"BUYER_COUNTY",
"TRANSACTION_DATE",
],
dtype={"TRANSACTION_DATE": "float64"},
)
df["year"] = df.TRANSACTION_DATE % 10_000
df["mme"] = df["MME_Conversion_Factor"] * df["CALC_BASE_WT_IN_GM"]
grouped = df.groupby(["BUYER_COUNTY", "BUYER_STATE", "year"])["mme"].sum().reset_index()
results = grouped.compute()
results.head()
| BUYER_COUNTY | BUYER_STATE | year | mme | |
|---|---|---|---|---|
| 0 | BOUNDARY | ID | 2011.0 | 4978.404375 |
| 1 | BRADFORD | PA | 2012.0 | 24691.530015 |
| 2 | BRISTOL | VA | 2012.0 | 17696.184008 |
| 3 | BROOMFIELD | CO | 2011.0 | 31757.599130 |
| 4 | COLUMBIA | OR | 2012.0 | 22251.313377 |
Exercise 6#
Now, re-write your opioid project’s initial opioid import using dask. Each person on your team should create a NEW branch to try this. The person who wrote the initial chunking code can help everyone else understand what they did originally and the data, but everyone should write their own code.
WARNING: You will probably run into a lot of type errors (depending on how the ARCOS data has changed since last year). With real world messy data one of the biggest problems with dask is that it struggles if halfway through dataset it discovers that the column it thought was floats contains text. That’s why, in the dask reading, I specified the column type for so many columns as objects explicitly. Then, because occasionally there data cleanliness issues, I had to do some converting data types by hand.
